Life, part 3
Code for this episode can be found here.
All right, let's get into it.
Since I want this series to concentrate on the algorithms and not the user interface, what I will probably do is make incremental improvements to the WinForms UI in each episode, but not discuss the details unless there is something particularly interesting.
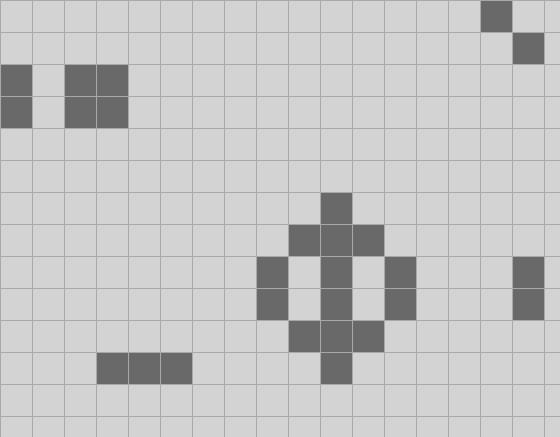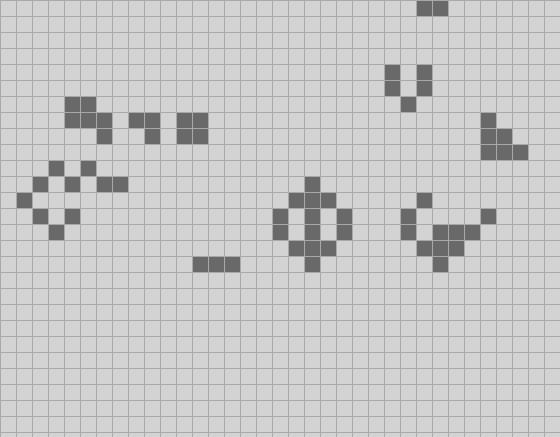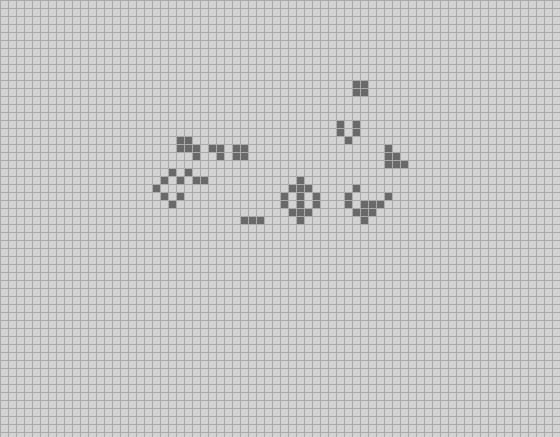
In the code for the previous episode the UI consisted just of a picture box that updated every second. The UI for this episode adds a new feature: you can now scroll with the mouse wheel to change the zoom level from cells being one pixel up to 64*64 pixels, in powers of two:
It's just like scrolling in a map application; the current position of the mouse determines the center point for the zoom.
I've also made the default starting configuration a Life form called acorn", because it is very small and grows into a surprisingly large tree. It takes over 5000 ticks to settle down into a stable configuration and throws off a bunch of gliders as it does. The screen shots above are from the early stages in acorn's evolution; check out the whole thing. It is quite mesmerizing. (Though you might want to adjust the timing in the code to compute more than two ticks per second!)
The first problem we face when designing a Life algorithm is the unavoidable fact that the board is in theory infinite, but our time and space available are finite. There are three basic ways to deal with this problem, and they all have their own pros and cons:
Fixed subset, dead everywhere else
Pick a board size: 256 x 256 cells, say. Everything outside the rectangle with corners (0, 0) and (255, 255) is considered permanently dead regardless of whether it really ought" to be alive. We pick a size that is big enough" for examining whatever Life form it is we wish to examine and do not worry about it further. If we end up with living cells near the border that would cause cells outside the border to come alive, we ignore that and they stay dead.
Fixed subset, wrap-around borders
Again, pick a finite size, treat everything outside that rectangle as permanently dead. But this time, make the board wrap around" such that the north" edge is adjacent to the south" edge and the east" edge is adjacent to the west" edge. If, say, a glider is heading towards the south edge, when it gets there it just keeps on going and appears on the north side of the rectangle.
There are two basic ways of conceptualizing this. The first and most common is that instead of an infinite flat grid, we've mapped a finite rectangular grid onto the surface of a torus - a doughnut. Imagine you took a rubber rectangle, rolled it into a cylinder to affix the north edge to the south edge, and then folded it to affix the now-circular west edge to the east edge. You'd end up with a torus.
The less common conceptualization is that we still have an infinite flat board, but we have tiled it with infinitely many copies of our rectangle; when we make a change to one tile, all the tiles change.
This technique is attractive from an implementation perspective because every cell has eight mutable neighbors and computing which cells are neighboring which others is simple math. However, it has a lot of problems too, particularly: if a Life form causes a glider to form that escapes", it ought to run off to infinity and bother no one again. But on a doughnut board it wraps around and ends up shooting straight at the cells that created it, possibly interfering with the evolution of the configuration.
We won't be using this technique in this series.
Embiggen as needed
We start with a fixed subset that is as large as we need, but when we have a cell that might come alive on the border, we push the boundary out. There are a lot of clever ways to do this; our final algorithm in this series will use this idea to some extent.
Enough chit-chat; let's look at the code for what I call the naive algorithm. The basic idea is: create a 256*256 array of bools; true means alive, false means dead. On every tick, create a new array, check every cell's neighbors in the old array to see if the cell will be alive in the next tick, and then discard the old array.
I'll omit the boring boilerplate code.
Initialization is straightforward; the default state of a bool array is all false:
class BoolArrayLife : ILife{ private int height = 256; private int width = 256; private bool[,] cells; public BoolArrayLife() { Clear(); } public void Clear() { cells = new bool[width, height]; }Everything outside our rectangle is always dead:
private bool IsValidPoint(long x, long y) => 0 <= x && x < width && 0 <= y && y < height; public bool this[long x, long y] { get { if (IsValidPoint(x, y)) return cells[x, y]; return false; }And similarly for the setter; you see how this goes.
To determine how many living neighbors a cell has, we just ask the indexer for the state of each neighbor. If the cell is out of range, the indexer tells us it is dead:
private int LivingNeighbors(int x, int y) { int sum = this[x - 1, y - 1] ? 1 : 0; sum += this[x - 1, y] ? 1 : 0; sum += this[x - 1, y + 1] ? 1 : 0; sum += this[x, y - 1] ? 1 : 0; sum += this[x, y + 1] ? 1 : 0; sum += this[x + 1, y - 1] ? 1 : 0; sum += this[x + 1, y] ? 1 : 0; sum += this[x + 1, y + 1] ? 1 : 0; return sum; }And the core inner loop" algorithm - that is, what do we do on each tick? - is as straightforward as can be:
public void Step() { bool[,] newCells = new bool[width, height]; for (int y = 0; y < height; y += 1) for (int x = 0; x < width; x += 1) { int count = LivingNeighbors(x, y); newCells[x, y] = count == 3 || (cells[x, y] && count == 2); } cells = newCells; }Recall that the client asks the engine to call it back for every living cell inside a certain rectangle. Were you ever asked the question how do you compute the intersection of two rectangles?" in an interview? It's a boring and cliched question but sometimes you actually do need to loop over the intersection of two rectangles!
public void Draw(LifeRect rect, Action<LifePoint> setPixel){ long xmin = Max(0, rect.X); long xmax = Min(width, rect.X + rect.Width); long ymin = Max(0, rect.Y - rect.Height + 1); long ymax = Min(height, rect.Y + 1); for (long y = ymin; y < ymax; y += 1) for (long x = xmin; x < xmax; x += 1) if (cells[x, y]) setPixel(new LifePoint(x, y));}And that's it! Just a bunch of loops.
Next time on FAIC: We'll start exploring some of the questions that I want to discuss in this series, like:
- What are the correctness and performance characteristics of this solution?
- Are there easy ways we could reduce the computational burden?
- How does it scale?
- What can we learn about optimization in general from optimizing to solve this problem in particular?