NYU Tandon Exploring “Megabase-Scale” Genetic Engineering

This is a sponsored article brought to you by NYU Tandon School of Engineering.
The human genome is built from 23 chromosomes. Within those chromosomes are around 3 billion base pairs of DNA. Within these base pairs are every subtlety of what makes you uniquely you - the way your eyes change color in different lighting, the sound of your laugh, your freckles.
It also encodes dangers. A host of genetic diseases and disorders can lurk among those 3 billion base pairs, in sequences that can cause multiple sclerosis, Alzheimer's disease, and more.
Since the completion of the Human Genome Project in 2003, scientists and researchers have pored over the map of our shared genetics to pick apart the clues and find the root causes of a host of human health problems. Their research has resulted in an explosion of knowledge that represents one of the great scientific advancements in human history. It also kickstarted the field of genetic engineering - the study of altering the genome to fight disease and change human health for the better.

With advancements like CRISPR, genetic engineering is entering its own renaissance. But while most geneticists are focusing on a few thousand base pairs at a time, some researchers are thinking bigger. Researchers like David Truong, Assistant Professor of Biomedical Engineering at the NYU Tandon School of Engineering and Associated Faculty of Pathology at the NYU School of Medicine. Truong is pushing the boundaries of biomedical engineering when it comes to genetics, building the technology necessary to change not just thousands of base pairs at a time, but millions. And that engineering work could have profound implications for the future of healthcare.
Truong is conducting his research with funding from a prestigious National Institute of Allergy and Infectious Diseases DP2 New Innovator Award, and his other laurels include a Delil Nasser Award for Professional Development from the Genetics Society of America, and a National Institutes of Health Ruth L. Kirschstein National Research Service Award.
We spoke with Truong about his work on genetics, and what it takes to work with huge amounts of genetic data at a time.
When did you become interested in genetic engineering?
When I was an undergraduate, I didn't actually major in bioengineering. I was specifically interested in manipulating the genome, but in the early 2000s, bioengineering wasn't that interested in human genomics. I pursued a molecular biology degree, but that was more of a means toward an end, which was the emerging field of genetic engineering.
Around that time, the Human Genome Project was finally published. Once we had the map of human DNA, it was on everyone's mind. And suddenly everyone understood that knowing the sequence of the human genome meant that we could start to manipulate that genome towards different ends, to cure diseases, to change medicine. At that point, bioengineering began to explode, and I was there to ride that wave of interest.
I did my Ph.D. at The University of Texas at Austin, producing a technique to directly edit genes. It was actually very similar to CRISPR-Cas9 - it combined a protein and a strand of RNA just like CRISPR. CRISPR came out right at the end of my Ph.D. studies, and unfortunately, I was never able to get my technology to work in human cells as well as the more well-known gene editing technique. So once I published my findings, like most genetic engineers, I moved over to CRISPR.
What brought you to NYU Tandon?
After my Ph.D. I was brought into the lab of Jef Boeke. He is a Professor of Biochemistry and Molecular Pharmacology at NYU Langone, as well as a Professor of Biomedical Engineering at Tandon. As a postdoc, I continued doing research into genomics.
The Boeke Lab is largely interested in the DNA of yeast and using that model as a way to explore synthetic genomics and building entirely new genomes. In his lab, yeast is used as a platform for exploring the construction of fully synthetic chromosomes.
But from that kind of cultural environment I started thinking about How do we manipulate the genome at this much larger scale?" - building larger sections of the human genome from scratch or modifying sections, or building whole chromosomes. So by the end of my post-doc work, I was already transitioning to working on what I had dreamed about as an undergraduate - building human cell therapies by using synthetic genomics and technologies.
I also helped during the early founding of a company while I was at the Boeke Lab called Neochromosome. I had written a small business grant to work on mammalian genome engineering, which would be a big focus of the company. It grew quite quickly once we had the funding, and it was acquired by Opentrons, who scaled Neochromosome up.
Eventually though, we realized that the market didn't have the patience for the type of work I do. The kind of large-scale genetic engineering I was pursuing was maybe 15 years away, and that's before clinical trials and regulatory approvals start. So they tracked back towards engineering yeast for more immediate gains, and I began to look back towards academia to really focus on the future of my work.
When a position in the Biomedical Engineering department opened up, I jumped at the chance. I was already familiar with NYU from my work with Boeke's lab, and I was eager to rejoin the community.
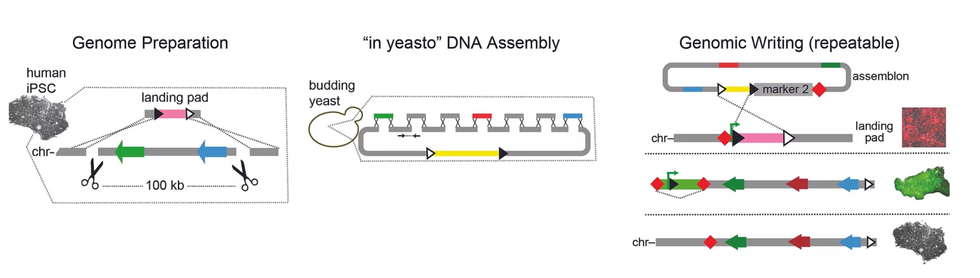
We had gotten pretty good at building designer segments of one hundred thousand base pairs, or even 1 million base pairs. That's what we call the mega-base scale ... It allows you to make huge adjustments to the way the genome acts and you can reimagine the chromosome in important ways."
-David Truong
When you say a larger scale," how large are we talking?
Most genetic engineers are focusing on small amounts of DNA - a portion of a gene, or a couple of nucleotides. Mostly looking to change a mutation here or there in a very limited capacity. That can involve five-to-ten thousand base pairs.
We had gotten pretty good at building designer segments of one hundred thousand base pairs, or even 1 million base pairs. That's what we call the mega-base scale, where you're changing many, many genes, as well as encoding the information that turns them off and on on-demand. It obviously allows you to make huge adjustments to the way the genome acts and you can reimagine the chromosome in important ways.
Doing this requires some specific technologies that we had to develop. CRISPR is our baseline, which we use to remove large sections of DNA. But in order to replace it, we use what we call landing pad" technology, which is optimized to receive these large pieces of genetic data.
And going back to my work in Professor Boeke's lab, we still use yeast to manufacture these large segments. Because most commercial DNA producers can't put such large strings of synthetic DNA together, we utilize yeast's unique abilities to take those small strands and combine them into something much larger than was previously possible. And those large chunks of DNA can be used in incredible ways.
A lot of your research focuses on human induced Pluripotent Stem Cells (iPSCs), which have the potential to transform from an embryonic state into any type of cell needed. What makes these cells so attractive to genetic engineering?
So iPSCs are cells typically taken from blood or skin, and reprogrammed to take on the properties of embryonic stem cells - those are the cells that can morph into the various kinds of cells that populate our body, from heart cells to brain cells to anything you can imagine. And because those cells have such remarkable plasticity, we can make these genetic adjustments to have them grow into cells with a very specific purpose.
One of the challenges, even with these cells, is that they can be highly personalized. So cells harvested from one person might be rejected by another. But what I've helped create was the technology to swap out the genetic coding in these cells that personalized them to an individual, and swap in genetic code that matches potential patients. And that is much cheaper and less invasive than having to start from scratch for each patient.
We're also engineering what we call synthetic genetic circuits. This is an idea in the synthetic biology field which takes from electrical engineering and computer science, where you actually use genes and the way they activate to actually power cells and change the way they act. The larger the amount of DNA you can swap out, the more complicated the function you can convince these cells to produce.
For example, one of the biggest things we're working on is turning iPSCs into cancer fighting cells. T-cells can hunt down tumors, but they are susceptible to the self-defense mechanisms of the tumors themselves, which also involves shutting down the body's natural immune system surrounding it. So with this technology, we can train" these cells to get around those suppression mechanisms and turn the immune system around the tumor back on, recruiting the body's natural defense system to help defeat the cancerous cells.
But we have a bunch of different potential uses for these cells. We're working with various kinds of immune cells. For example, dendritic cells move around the body, picking up bits of protein and other molecules that they find, and when they find something that's not supposed to be there, they put it on their surface. And then T-cells can learn' from these molecules what they should be attacking in the body. So we can harvest these T-cells from patients that are adapted to attack certain types of cancer, and recreate synthetic T-cells that can be introduced to new patients to kick-start their body's defense system.
How does your work dovetail with others at NYU Tandon?
There's a lot of opportunity for collaboration in the NYU Tandon BME department [Editor's note: The department was covered in the pages of this website earlier this year]. Irene de Lazaro, the newest Assistant Professor of Biomedical Engineering, also works with iPSCs. She's doing very similar work, using these stem cells to produce therapies, specifically rejuvenating heart cells. There's a lot of complementary overlap there. Alesha Castillo is working on stem cells, and Thorsten Kirsch is working on how cellular interactions affect things like osteoarthritis. So cell and tissue engineering is becoming an ever-growing field of study at the school, and that makes it a very exciting place to be right now.
Then there's research like Weiqiang Chen's, who is currently working on cancer-on-a-chip technology. Essentially, he's producing miniature, personalized cancer samples that represent an individual's tumor. Theoretically, if he was looking to test a very specific treatment involving specialized genetically-modified cells, he could come to our lab and we could produce them for him, and quickly test in a controlled sample whether a treatment had the potential to work for any given patient.
That's what really excites me about this work. As bioengineering grows, it's interfacing more with medicine, and we're getting to the place where we can use cells and genomes as a technology itself. When I was thinking about bioengineering as an undergraduate, it wasn't a thing. It's only now that we have learned enough about nature, enough about the rules, that we can really start to take these things and put them together in new ways for new medical treatments and human health in general. I think it's just a huge opportunity to grow.