Getting software to “hallucinate” reasonable protein structures
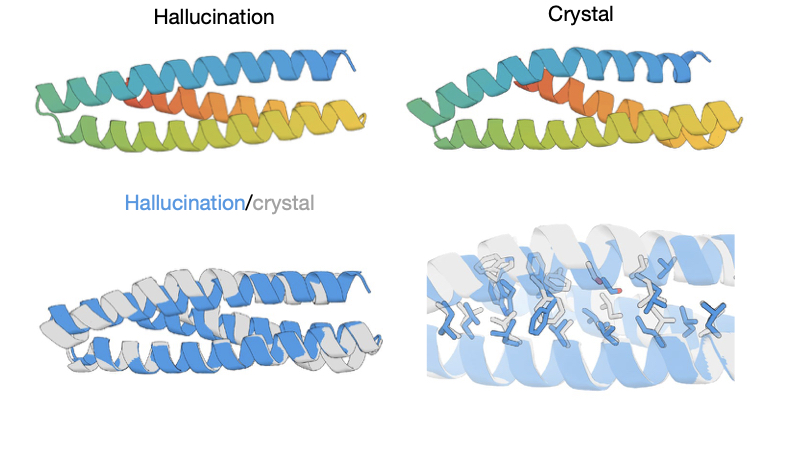
Enlarge / Top row: the hallucination and actual structure. Bottom row: the two structures superimposed. (credit: Anishchenko et. al.)
Chemically, proteins are just a long string of amino acids. Their amazing properties come about because that chain can fold up into a complex, three-dimensional shape. So understanding the rules that govern this folding could not only give us insights into the proteins that life uses but could potentially help us design new proteins with novel chemical abilities.
There's been remarkable progress on the first half of that problem recently: researchers have tuned AIs to sort through the evolutionary relationships among proteins and relate common features to structures. As of yet, however, those algorithms aren't any help for designing new proteins from scratch. But that may change, thanks to the methods described in a paper released on Wednesday.
In it, a large team of researchers describes what it terms protein "hallucinations." These are the products of a process that resembles a game of hotter/colder with an algorithm, starting with a random sequence of amino acids, making a change, and asking, "Does this look more or less like a structured protein?" Several of the results were tested and do, in fact, fold up like they were predicted to.
Read 14 remaining paragraphs | Comments